
An individual who is heterozygous for a gene has two different alleles, but it is not always sufficient to produce an intermediate phenotype. All progeny were pink flowered, an intermediate phenotype. Had he worked with Mirabilis jalapa rather than pea he would have pondered the cross of a plant with red flowers by one with white flowers. Mendel described dominance but not incomplete dominance. Incomplete dominance is also referred to as semi-dominance and partial dominance.
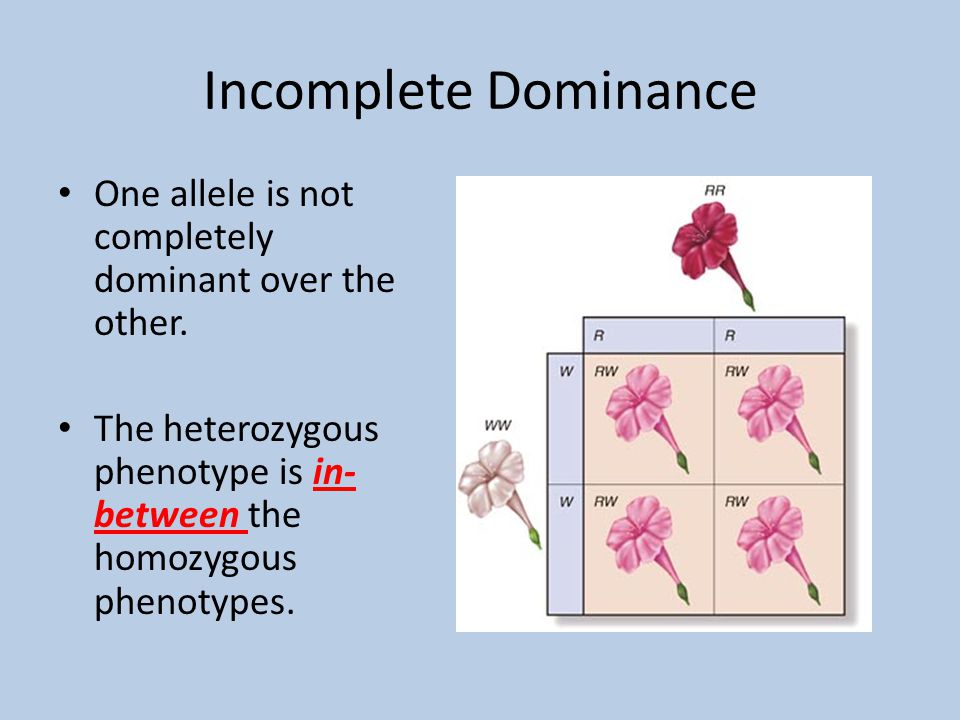
Incomplete dominance results from a cross in which each parental contribution is genetically unique and gives rise to progeny whose phenotype is intermediate. Frizzell, in Brenner's Encyclopedia of Genetics (Second Edition), 2013 Abstract Our results provide empirical support for an important role for incomplete dominance of deleterious alleles in explaining heterosis and demonstrate the utility of incorporating functional annotation in phenotypic prediction and plant breeding.M.A. Cross-validation results show that incorporating sequence conservation in genomic selection improves prediction accuracy for grain yield and other fitness-related traits as well as heterosis for those traits. We identify haplotype blocks using an identity-by-decent (IBD) analysis and perform genomic prediction analyses in which we weigh blocks on the basis of complementation for segregating putatively deleterious variants. Our modeling reveals widespread evidence for incomplete dominance at these loci, and supports theoretical models that more damaging variants are usually more recessive. Inbred elite maize lines vary for more than 350,000 putatively deleterious sites, but show a lower burden of such sites than a comparable set of traditional landraces.

We measured a number of agronomic traits in both the inbred parents and hybrids of an elite maize partial diallel population and re-sequenced the parents of the population. We test these ideas using evolutionary measures of sequence conservation to ask whether incorporating information about putatively deleterious alleles can inform genomic selection (GS) models and improve phenotypic prediction.
:max_bytes(150000):strip_icc()/incomplete_vs_codominance2-2e6704aac494409fa555975e560cab4e.jpg)
2 DuPont Pioneer, Johnston, Iowa, United States of America.

1 Department of Plant Sciences, University of California, Davis, Davis, California, United States of America.


 0 kommentar(er)
0 kommentar(er)
